Introduction:

Dr Sarah Leiter is currently a ST3 paediatric trainee and Academic Clinical Fellow (ACF) in the East of England, with an interest in paediatric oncology. She studied Medicine at the University of Cambridge and did an MB/PhD studying the genetics and mechanism of an ultrarare form of neonatal hypoglycaemia.
Outside of research and being a clinician, she is heavily involved in the Paralympic sport goalball, and only recently retired from representing Great Britain internationally!

Sarah is the joint first author for a recent Nature Medicine publication, which was covered by many media sources, including the BBC (articles linked below).

Can you tell us about the study – what motivated the research, and what were the key messages?
Previously, whole genome sequencing (WGS) in children with cancer wasn’t done routinely – it would generally be done on high risk populations, such as children with difficult to treat cancers, or when cancers relapse, or to investigate the genomic features of the cancer as part of research projects. This study is novel as it looks for the clinical impact that is made at the time that routine WGS is offered to every child with cancer, on their care, in real time.
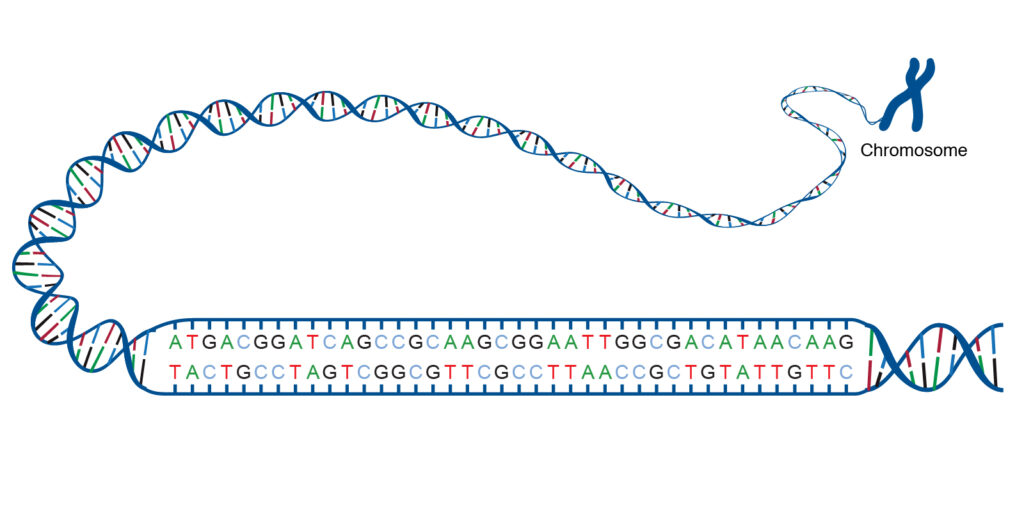
Whole Genome Sequencing is a method to read the order of DNA bases (A, G, C, T) of someone’s entire genome, which can be compared against a reference genome to identify mutations, or base changes, which may be contributing to a disease or condition. (image from https://www.genome.gov/genetics-glossary/acgt)
The key finding is that in 7% of children, WGS changed the management of the child which wouldn’t have happened through other means of investigation. By sequencing both the tumour and germline DNA we were able to find unexpected predisposition syndromes in some patients ensuring they receive additional surveillance and altered treatments as indicated. In other children we re-defined their tumour type optimising their treatments and thus likely their long-term outcomes. We are fortunate that NHS England has commissioned WGS for every young person under the age of 25 with suspected cancer as part of their routine clinical care. Our research demonstrates the real-time real-life impact that routine WGS can have for these children.
What was the reality like to work on such a massive project?
I had nine months of full-time research time as part of the ACF, which gave me the space to truly focus on the project and do a large bulk of the work, though naturally I continued working on it after my return to the clinical work. Balancing academic and clinical careers is challenging but also really rewarding. I worked in close collaboration with both the local clinical teams in Cambridge and the haematology team at GOSH in London. Working as a large team means you can create more impactful research but can provide challenges and requires good communication between all those involved.

Left: at the Children’s Cancer and Leukaemia Group Conference 2024, Sarah received a Highly Commended Research Award for this project
Paediatric cancers are considered rare diseases – what is the significance of this?
Paediatric cancers are rare as they only affect a small number of children with an estimated 1900 new diagnoses in the UK each year. There are many different types of cancers and whilst some have been well studied for decades relatively little is know about others. In addition to providing a real-life clinical impact to children with cancer, WGS will provide us with many valuable insights into these rare tumours.
As someone who has had a heavy focus on research throughout your medical school and specialty training, how has your experiences and motivations for research changed or developed over time?
I was interested in doing the MB/PhD programme as I knew I wanted to do research from a young age. Doing my PhD early on during medical school really suited me and I loved it. I like to understand how things work at a very detailed level, and this helps me to understand the background to clinical decisions that we make. My motivation for research has always been to optimise the care and outcomes for the patients we treat.
My PhD was a mostly wet lab project working on creating in vitro models of rare genetic conditions. I did a lot s sequencing and cell culture as well as assay development. I enjoyed working in the lab as it gave me a very good foundation in research.
I didn’t do an academic foundation programme, but maintained my involvement in research during this time through getting involved in small clinical projects and finishing off some writing from my PhD.
This project opportunity during my ACF really interested me, as it was a clinically based study directly investigating how genomics (my skill set from my PhD) can have a real-life impact on patients. I had to newly learn a variety of bioinformatics skills, such as UNIX and programming using R. It was a steep learning curve, but also very exciting and rewarding.
I am planning to pursue paediatric oncology training and continue with genomics research using everything that I have already learnt and learning many new things along the way!
Additional links:
BBC news article: https://www.bbc.co.uk/news/articles/crgk4r308mro
Cambridge University Hospitals article: https://www.cuh.nhs.uk/news/cutting-edge-genomic-test-can-improve-care-of-children-with-cancer/#:~:text=Overall%2C
Nature Medicine original publication: https://www.nature.com/articles/s41591-024-03056-w
